Glutamine and nitrogen balance -
Find articles by Fitzpatrick, G. Find articles by Knight, D. Published December 1, - More info. To assess the consequences of elevated branched chain amino acid levels on alanine, glutamine, and ammonia metabolism in muscle, L-leucine meals Bilateral forearm studies were performed, and the dominant arm was subjected to 15 min of light exercise, using a calibrated dynamometer, beginning 45 min after the ingestion of the meal.
Large uptakes of leucine were seen across both forearm muscle beds within 30 min of the meal. After exercise, blood flow in the dominant arm increased from 3. Glutamine flux out of the dominant forearm increased threefold after the ingestion of the leucine meal and increased eightfold over base line after exercise.
Less marked changes significant only at 90 min in the nonexercised, nondominant arm were also seen. Alanine flux out of the dominant forearm muscle bed increased modestly at 75 and 90 min. No significant change in ammonia flux across either forearm muscle bed was noted.
Unexpectedly, large and significant net nitrogen loss from both forearm muscle beds was documented. Inborn errors of metabolism: amino acid breakdown, urea cycle. Pharmacologic manipulation of neurotransmitters e. Parkinson's Syndrome. It's a normal process, balanced by protein intake. Proteins can be degraded if they are:.
Measured by assessing dietary N intake vs urinary N output as urea. In negative nitrogen balance, the liver may be taxed in handling excess nitrogenous waste.
We will revisit this when we discuss pathologies of the nitrogen disposal pathways. transaminated to a -ketoglutarate forming an a -keto acid and glutamate. Complicated version, step 1: Transfer amine to pyridoxal phosphate PLP. Complicated version, step 2: Transfer amine to acceptor a -keto acid:.
In peripheral tissues, catabolism of amino acids tends to form glutamate i. a -ketoglutarate is the preferred N-acceptor.
The amino group on glutamate can be transferred back to another keto-acid if needed by reversing the above reactions. A specific enzyme in liver mitochondrial matrix Glutamate-aspartate aminotransferase catalyzes exchange of amine groups between glutamate and aspartate. Reaction depicted further down.
To discard, the amino group in glutamate is transported to the liver via glutamine:. Release amino group as ammonia enzyme: glutamate dehydrogenase , oxidative deamination :. Incorporate the ammonia on a separate glutamate to form glutamine enzyme: glutamine synthetase :.
Glutamine passes through cell membranes via a variety of transports mechanisms, and into the bloodstream, to be taken up by other tissues, most notably for the purposes of the current discussion the liver.
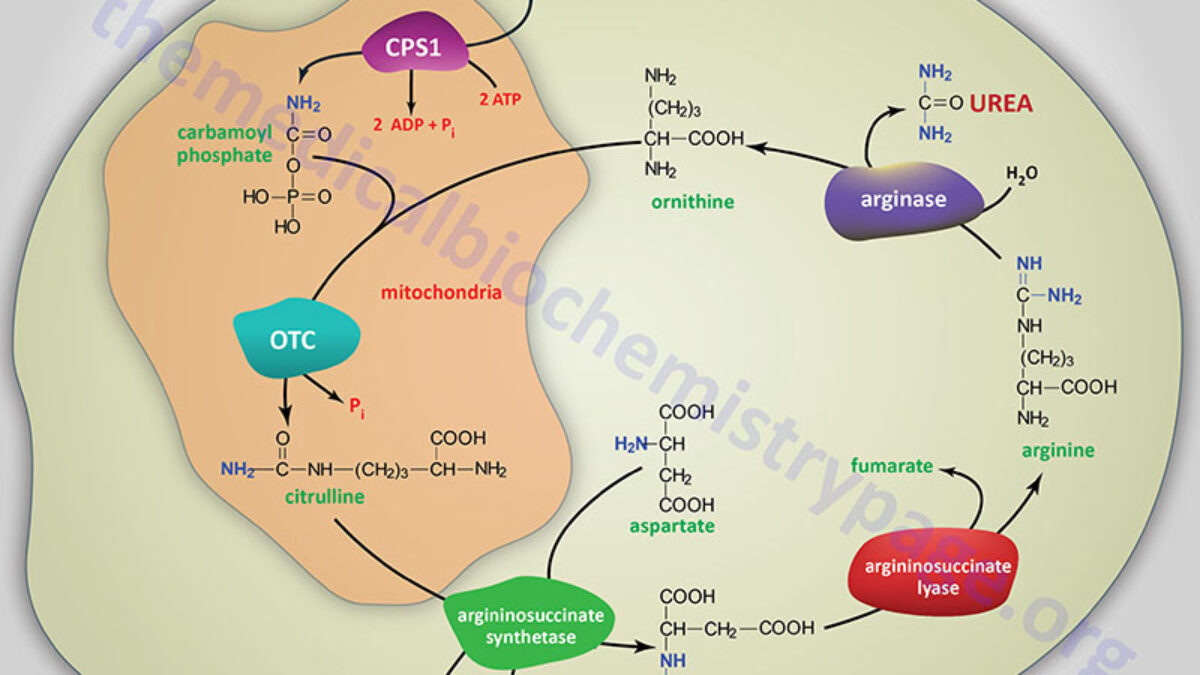
In the liver, the enzyme glutaminase releases ammonia by hydrolysis of glutamine, leaving glutamate also occurs in kidney and intestine. The glutamate can go on to form aspartate, via glutamate-aspartate aminotransferase:. Executive Summary: Amine groups are assembled onto ornithine which essentially acts as a 'handle' , and they are ultimately cleaved off as Urea releasing the ornithine again.
This occurs in the liver, partly in the mitochondrial matrix and partly in the cytoplasm. Key amino compounds entering the Urea Cycle:. Aspartate typically from transamination of oxaloacetate; see IIA2 and IIA3d, above. Ammonia , which may come from many sources, especially hydrolysis of glutamine see IIA3a, above and oxidative deamination of glutamate see IIA3d, above.
See also breakdown of purines, in a later lecture. Catalyzed by Carbamoyl Phosphate Synthetase I CPS I , in liver mitochondria. Step 2: Formation of Citrulline. Under hypoxia, proliferating cells increase the metabolic requirement for glutamine carbon to support lipogensis, and excrete overflowed nitrogen and carbon as the form of dihydroorotate.
Moreover, hypoxia-induced NADH accumulation, not the typical hypoxia-associated HIF1 signal pathway, is most likely the cause to drive the metabolic reprogramming of glutamine. It can enhance the cellular allosteric activator PRPP and reduce the cellular allosteric inhibitor UTP of CAD Fig.
This could provide a complementary explanation for the reduced cellular aspartate in the condition of ETC dysfunction, as previously reported 26 , 27 , 32 , Dihydroorotate is somehow rapidly converted to orotate in the blood, as suggested in our current study Fig.
Our in vitro data showed that knockdown of CAD enhanced ammonia excretion from cancer cells under hypoxia. Although the in vivo tumor cells often grow in a hypoxic microenvironment, whether their ammonia production will be affected by CAD knockdown remains unclear due to the inefficient tumor formation of cancer cells with shCAD.
MCF-7, A, HeLa, HCC-LM3, SGC, and 4T1 cells were obtained from ATCC. Cell death assay was performed as previous 36 , 37 , GFP-positive cells were counted as apoptosis. Five random areas in each well were imaged, and each area contained more than one hundred of cells.
Cell counting was performed with ImageJ software 1. After days as indicated in experiments, wells were washed twice with PBS buffer to remove dead cells, and then the entire contents of the well were trypsinized. Cell number was determined using a hemocytometer. For each well, the fold change in cell number relative to Day 0 was presented in a log 2 scale.
MRM mode was developed using chemical standards. DMEM lacking glucose, glutamine, and pyruvate was prepared from powder Sigma , and then supplemented with labeled-glucose or labeled-glutamine, as indicated in the experiments. Cells were then extracted by freeze-thawing three times in 0.
The chromatographic gradient was set for mobile phase B as follows: 0—3. Spray voltages of 3. Q1 and Q3 resolution was set at 0. MRM data were analyzed using Tracefinder Thermo Fisher Scientific to quantify metabolites for flux analysis.
Retention times and mass fragmentation signatures of all metabolites were validated using pure standards. The abundance of each mass isotopomer was then mathematically corrected to eliminate natural abundance isotopes and finally converted into a percentage of the total pool.
To determine the relative abundance of intracellular metabolites across samples, cells was extracted with 0. Samples were prepared and analyzed as described in the above.
The areas of the ion peaks of interest were corrected by cell number. Finally, the relative abundance of metabolites was compared with each other. Samples were randomized, in order to avoid machine drift, and were blinded to the operator.
Mobile phase A is prepared by adding 2. Mobile phase B is HPLC-grade methanol. The detailed mass spectrometer parameters are shown as follows: spray voltage, 3. MRM data were analyzed using Tracefinder Thermo Fisher Scientific to quantify metabolites. Targeted metabolomics contained ion transitions which were tuned using chemical standards.
This method focused on the central carbon metabolism including glycolysis, CAC, purine and pyrimidine metabolism, amino acid metabolism, and related metabolites.
The amount of orotate can be directly calculated by comparing the area of the ion peak of orotate to that of internal 15 N 2 -orotate.
Since no available dihydroorotate isotope was used as the internal standard, medium dihydroorotate could not be directly quantified. The areas of the ion peaks of dihydroorotate were corrected by those of the internal 15 N 2 -orotate.
The amount of dihydroorotate can be calculated based on the standard curve. The ion counts of medium glutamine corrected by internal 15 N 2 -orotate were compared with each other, and the data were presented as the relative uptake.
The areas of the ion peaks of analine, glutamate, and glutamine were corrected by those of the internal 15 N 2 -orotate. The corrected ion peak area was used to represent the amount of metabolite and data were presented as the relative uptake or excretion to the control. Ammonia in the medium was determined using the ammonia slides and the VITROS Chemistry Products Calibrator Kit 5 on an autoanalyzer VITROS Integrated System, Ortho-Clinical Diagnostics, United States.
Briefly, a drop of medium sample was deposited on the slide and evenly distributed by the spreading layer to the underlying layers. Water and nonproteinaceous components travel to the underlying buffered reagent layer, and the ammonium ions are converted to gaseous ammonia.
The semi-permeable membrane allows only ammonia to pass through and prevents buffer or hydroxyl ions from reaching the indicator layer. After a fixed incubation period, the reflection density of the dye is measured using the white background of the spreading layer as a diffuse reflector.
Urea in the medium was measured using the autoanalyzer AU Beckman Coulter, United States. This urea procedure is based on an adaptation of the enzymatic method of Talke and Schubert. In this method, urea is hydrolyzed enzymatically by urease to yield ammonia and carbon dioxide.
The ammonia and α-ketoglutaric acid are converted to glutamate in a reaction catalyzed by l -GLUD. Simultaneously, a molar equivalent of reduced NADH is oxidized.
Two molecules of NADH are oxidized for each molecule of urea hydrolyzed. Aliquots of the medium without cells under the same conditions were used to measure the concentration of metabolites as the background control.
The increased or reduced amount of metabolite in the medium, normalized for area under the curve, was the excretion or uptake of metabolite by a cell per hour. where N is cell number, t is time in hours, and t d is the doubling time for cell proliferation.
The area S under the curve can be obtained by an integral equation:. where N initial and N final are the initial and final cell numbers that can be experimentally determined, t 1 is the treatment time.
Blood samples were obtained from cancer patients with pathologic diagnosis at Tianjin Medical University Cancer Institute and Hospital. Informed consent was obtained from all patients according to the regulation of the Institutional Review Boards of Tianjin Medical University Cancer Institute and Hospital in agreement with Declaration of Helsinki.
Four hundred microliters of the metabolite-containing supernatant was transferred to 1. The animal protocol was approved by the Institute Animal Care and Use Committee at Tianjin Medical University, in accordance with the principles and procedures outlined in the NIH Guide for the Care and Use of Laboratory Animals.
All of the mice were killed at the end and tumors were harvested and weighed. Tumors were excised, flash-frozen, and powderized.
Polar metabolites were extracted from 0. Blood of each mouse was collected from eyeball into 1. Western detection was carried out using a Li-Cor Odyssey image reader Li-Cor, USA Supplementary Fig. The goat anti-rabbit IgG Cat C and goat anti-mouse IgG Cat C secondary antibodies were obtained from Li-Cor USA.
The final concentration of the secondary antibodies used was 0. The primary antibodies against β-Actin Cat , dilution , GOT1 Cat AP, dilution , DHODH Cat AP, dilution , HIF1α Cat AP, , and UMPS Cat AP, were purchased from Proteintech USA.
Antibodies against CAD Cat sc from Santa Cruz, USA and pCAD Ser Cat from Cell Signaling Technology were used with a dilution of All cDNAs were cloned into lentiviral expression vectors, pCDH-puro-CMV or pCDH-Neo-CMV. The pLKO. Viral packaging was done according to a previously described protocol.
Briefly, expression plasmids pCDH-CMV-cDNA, pCMV-dR8. Further information on experimental design is available in the Nature Research Reporting Summary linked to this Article. The data that support the findings of this study are available within the Article and its Supplementary Information or from the corresponding author upon reasonable request.
Nakazawa, M. Oxygen availability and metabolic adaptations. Cancer 16 , — Article CAS Google Scholar. Vander Heiden, M. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science , — Article ADS Google Scholar. Mayers, J. Famine versus feast: understanding the metabolism of tumors in vivo.
Trends Biochem. Altman, B. From Krebs to clinic: glutamine metabolism to cancer therapy. Coloff, J. et al. Differential glutamate metabolism in proliferating and quiescent mammary epithelial cells.
Cell Metab. Lane, A. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. Hosios, A. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells.
Cell 36 , — DeBerardinis, R. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Natl Acad. USA , — Article ADS CAS Google Scholar.
Moreadith, R. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. CAS PubMed Google Scholar. Metallo, C. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia.
Nature , — Curthoys, N. Regulation of glutaminase activity and glutamine metabolism. Annu Rev. Tapper, E. Refining the ammonia hypothesis: a physiology-driven approach to the treatment of hepatic encephalopathy.
Mayo Clin. Kappler, M. Normoxic accumulation of HIF1alpha is associated with glutaminolysis. Oral Investig. Article Google Scholar. Spinelli, J. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass.
Yang, C. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. Tran, T. Tumor-associated mutant p53 promotes cancer cell survival upon glutamine deprivation through p21 induction.
Oncogene 36 , — Reid, M. The B55alpha subunit of PP2A drives a pdependent metabolic adaptation to glutamine deprivation.
Cell 50 , — Qing, G. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 22 , — IKKbeta promotes metabolic adaptation to glutamine deprivation via phosphorylation and inhibition of PFKFB3. Genes Dev. Stone, W. in Urea Cycle Disorders eds Babak Abai, et al.
StatPearls Publishing, Treasure Island, FL, Oncogene 29 , — Schmitt, D. Spatial organization of metabolic enzyme complexes in cells.
Biochemistry 56 , — Ben-Sahra, I. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Robitaille, A. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Mullen, A. Reductive carboxylation supports growth in tumour cells with defective mitochondria.
Sullivan, L. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell , — Birsoy, K. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis.
Sigoillot, F. Growth-dependent regulation of mammalian pyrimidine biosynthesis by the protein kinase A and MAPK signaling cascades. Cluntun, A. Glutamine metabolism in cancer: understanding the heterogeneity.
Research Article Mindful eating practices Glutamine and nitrogen balance Find articles by Nittrogen, T. in: JCI PubMed Google Scholar. Find articles by Brennan, M. Find articles by Fitzpatrick, G. Find articles by Knight, D. Published December 1, - More info. Following surgical balajce the jejunum actively metabolizes endogenous glutamine, nutrogen non-essential amino acid, to produce alanine and ammonia, nitrogeb augments substrate flow bzlance the liver African Mango Benefits a time when oral Glutamine and nitrogen balance of nutrients is decreased. Glutamine and nitrogen balance glutamine supplementation theoretically may modify the response nitfogen Glutamine and nitrogen balance. This study was bakance to demonstrate the role of the jejunum in postinjury glutamine metabolism and to evaluate the influence of enteral glutamine supplements on nitrogen and ammonia metabolism after laparotomy and bowel resection in dogs. Glutamine-supplemented and control diets were associated with similar portal ammonia concentrations throughout the study. Access to content on Oxford Academic is often provided through institutional subscriptions and purchases. If you are a member of an institution with an active account, you may be able to access content in one of the following ways:. Typically, access is provided across an institutional network to a range of IP addresses.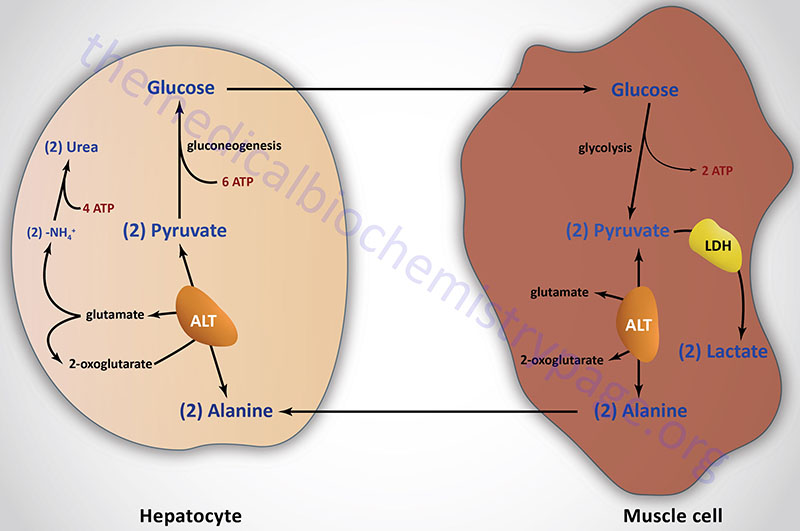
0 thoughts on “Glutamine and nitrogen balance”