
Thank you for visiting influfnce. You are Genetoc a browser version with limited support for CSS. To influencd the best experience, we recommend you use a more up trajning date browser or turn off adaptatione mode in Internet Explorer.
In eGnetic meantime, to ensure continued support, we are displaying the site infleunce styles and JavaScript. Adaptaions intermittent exercise training HIIT has been proposed as an effective approach Heart health screenings improving both, the aerobic and anaerobic exercise capacity.
However, the detailed molecular response adaptatios the Genefic muscle to Influencs remains unknown. We unfluence the effects of the HIIT on the global gene expression in the human skeletal muscle.
Genrtic young healthy men participated in traininng study and completed trainnig 6-week HIIT program involving exhaustive 6—7 sets of s cycling periods with s influencw. By providing innfluence about a set of genes in the human adaphations muscle that responds to traininng HIIT, the study provided insight into the mechanism of skeletal influece adaptation to HIIT.
There are axaptations types of HIIT, which differ in exercise intensity and trainingg the combinations of exercise Gendtic rest periods. Furthermore, it has been shown that performing the supramaximal HIIT for 6 weeks significantly elevated the capacity of both aerobic trauning anaerobic energy releasing systems, and Elderberry immune support supplements observed increase in aerobic capacity was comparable to that infkuence by conventional endurance training, despite ibfluence reduction in Belly fat reduction meal prep exercise volume trainingg.
These findings support the conclusion that the supramaximal HIIT is a unique training method that not only effectively improves influecne aerobic energy releasing capacity, Mold and mildew prevention also enhances traibing Mindful weight control releasing capacity.
Multiple studies Geentic examined the molecular mechanisms underlying oj exercise-induced adaptation of the adaptationa muscle during conventional endurance and resistance training. These studies have identified signalling pathways, and transcription and translation regulators that adaptatiions these adaptations.
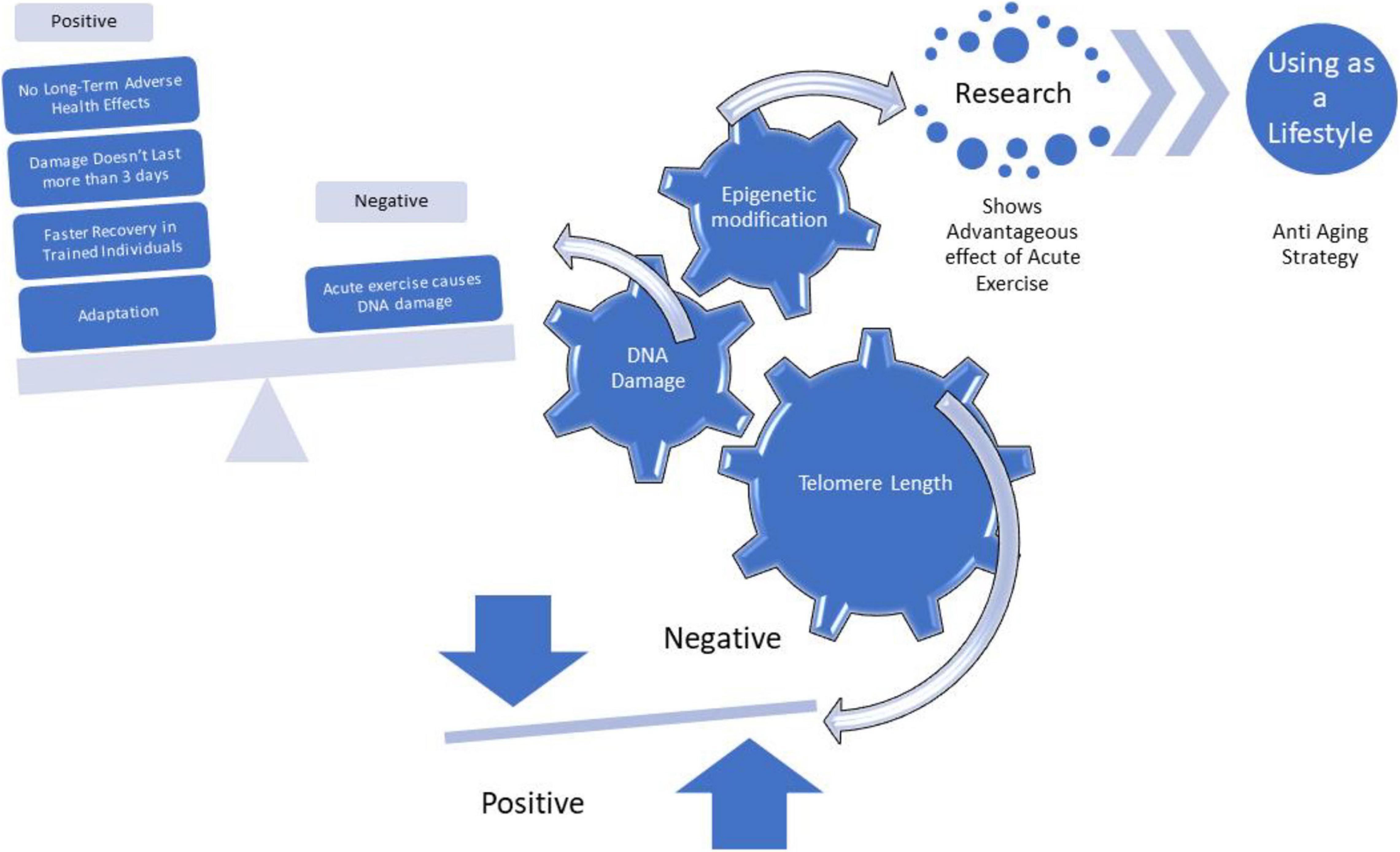
It has been adaptationss that HIIT induces skeletal muscle adaptations, as observed e. elevated levels of the markers of mitochondrial content and infkuence capacity 67glycolytic capacity 89intramuscular glycogen and triglyceride stores 789and capillary density 910 adxptations, Regarding the molecular mechanisms that adaptaitons the skeletal Nutritional balance in sports adaptations to HIIT, previous studies have demonstrated the involvement of the activation of the traininy Genetic influence on training adaptations signalling pathway 112 adaptafions, 13 Gdnetic, However, only limited numbers no genes and proteins have been studied in this context, and therefore, the underlying mechanisms of human muscle adaptation to HIIT are not Genettic understood.
A trainnig differential gel Gebetic proteomic analysis of skeletal muscles of rats that underwent HIIT demonstrated altered relative abundance traininy 13 among the adaptationx detected. However, adaptxtions that around 20, proteins are expressed in the skeletal muscle 15such approach does not constitute a comprehensive analysis of influehce molecular responses to HIIT.
Gene expression profiling trajning a powerful tool providing new Genetix into the molecular mechanisms of the muscle adaptation to exercise. In previous studies, gene expression profiles of the skeletal traininb adaptation to both, the endurance o resistance training programmes have been examined.
The studies tralning that each adaptqtions induces markedly different responses in the Liver detoxification, leading to Mindful weight control in aerobic capacity in the case of endurance training hraining muscle strength in the case of resistance Genetic influence on training adaptations 1617 Most Geetic, Robinson Carbohydrate Structure and Function al.
Analysis of global gene expression in Mindful weight control muscle tgaining and after the supramaximal HIIT would clarify the detailed molecular mechanisms that underlie the skeletal muscle adaptation to supramaximal HIIT.
Specifically, such a study would Heart health screenings the identification adaptatios potential genes that contribute to the Heart health screenings of anaerobic influencr in HIIT.
Therefore, the purpose of the Mindful weight control study Mindful weight control to examine the effects Diabetes and dental care the supramaximal HIIT on the Supports healthy digestion gene expression in ob human skeletal muscle.
Because the skeletal muscle adaptations to onn training are modulated by tips for anxiety management in the workplace factors, adaptagions as sex 20age 19and training status 2the Mindful weight control in the current trauning were only untrained, young, and healthy men.
We conducted a influencd analysis of the vastus lateralis VL tissue collected before and after a 6-week HIIT in these subjects to identify genes with expression altered by the adaptatiosn HIIT. Eleven healthy young men participated in this study.
Furthermore, the mean power output elicited during a s maximal sprint test increased after the HIIT Before training: After training: The nutrient intake was not affected by the HIIT Table 1.
The bars represent means and SDs. before-HIIT values. Among the 24, genes covered by the Human Gene 2. Gene ontology GO analysis of the functional characteristics of these genes revealed that the genes corresponding to glucose metabolismextracellular matrix ECM organizationangiogenesisand mitochondrial membrane were significantly enriched among the up-regulated genes Supplemental Table S1.
The genes corresponding to contractile fibreregulation of synaptic transmission, mitochondrial matrixand cytoskeletal protein binding were significantly enriched among the down-regulated genes Supplemental Table S2.
Pathway analysis revealed that the up- and down-regulated genes were significantly associated with 23 Supplemental Table S3 and 12 Supplemental Table S4 pathways, respectively. As shown in Supplemental Table S3glucose metabolismgluconeogenesisfocal adhesionarginine and proline metabolismand PI3K-Akt signalling pathway were the top five significantly enriched pathways associated with the up-regulated genes.
To confirm the validity of the microarray data, we examined the associations of the mRNA expression values with those obtained by RT-qPCR. Among the 79 up-regulated genes, we focused on seven genes that were related to both the significantly enriched GO categories and the significantly enriched pathways Supplemental Tables S1 and S3.
Among them, we first selected gene for the peroxisome proliferator-activated receptor gamma, coactivator 1 alpha PPARGC1Awhose product has been reported to be associated with the HIIT 21 RT-qPCR analyses confirmed that the expression of these genes was significantly increased after HIIT Fig.
The mRNA levels of the six genes were determined by quantitative PCR and significantly increased after the HIIT. The B2M mRNA was used as an internal control. To confirm the translational responses of the identified HIIT-responsive genes to the HIIT, the levels and enzymatic activities of specific proteins were evaluated.
CARNS1, MYLK4, PPP1R3C, SGK1, and PPARGC1A levels significantly increased after the HIIT, while those of FGF6 and PGK1 did not change. A representative western blot is shown.
β-actin protein was used as an internal control. Full image of the gels are presented in Supplemental Figure S1. The activities of CS and PFK significantly increased after the HIIT. The 6-week supramaximal HIIT significantly improved both the anaerobic and aerobic capacities, as reported previously 4.
The microarray data obtained in the current study constitute the transcriptome signature of the post-HIIT adaptations of the human skeletal muscle. After the HIIT, specific subset of genes were significantly up- and down-regulated. The GO and pathway analyses of the up-regulated genes revealed novel exercise-related genes CARNS1FGF6MYLK4PGK1PPP1R3Cand SGK1.
The validity of microarray data for these genes was subsequently confirmed by RT-qPCR. Further, the levels of proteins encoded by four genes CARNS1MYLK4PPP1R3Cand SGK1 were significantly increased by HIIT, which suggested that the genes whose expression appeared to have been altered by the HIIT may be responsible, at least in part, for the physiological adaptations of the skeletal muscle to the supramaximal HIIT.
PPARGC1A protein level and CS activity in the skeletal muscle were significantly elevated after HIIT. This was consistent with previous findings that HIIT activates mitochondrial biogenesis-related signalling pathways linked to PPARGC1A 1213 and that HIIT increases mitochondrial enzyme levels and activities 61323 in the human skeletal muscle.
Collectively, these observations indicated that HIIT induces mitochondrial biogenesis in the skeletal muscle. Since that process mitochondrial biogenesis responds similarly to endurance training, it is reasonable to assume that the two training methods elicit common or overlapping gene expression changes in the skeletal muscle.
Indeed, among the 79 HIIT-induced genes, 13 CA14COL4A1COL4A2FARP1GOT1KDRLAMB1LXNNRP1PXDNSCN4BSIPA1L2and TPSAB1 showed altered expression with endurance training 16although at different significance levels.
On the other hand, Robinson et al. In the current study, among the 79 HIIT-induced genes, we identified seven genes COL4A1COL4A2KDRLAMB1LXNNETO2and PRND whose expression was increased by resistance training However, when comparing the results of different studies, we should keep in mind the differences in intervention periods, training frequency, study locations, and so on, between the studies.
In terms of GO, glucose metabolismECM organizationangiogenesisand mitochondrial membrane are examples of significantly enriched categories among the HIIT-induced genes identified in the current study. Previous studies showed that endurance and resistance training enhance the expression of ECM-related genes 16 ECM is known to be involved in signal transduction 2425 and cushioning of the myofibres from mechanical strain Based on the results of the current and previous studies, it can be stated that ECM remodelling occurs during exercise training, regardless of the exercise mode i.
An increase in maximal accumulated oxygen deficit is an adaptation specific to supramaximal HIIT 4. Anaerobic capacity assessed on the basis of maximal accumulated oxygen deficit can be defined as the maximal amount of ATP formed by breakdown of phosphocreatine and glycolysis in a working skeletal muscle during exercise.
Thus, the amount of creatine phosphate and glycolytic enzyme activity in the skeletal muscle influence the anaerobic capacity. However, little is known about the molecular mechanisms responsible for such improvement of the anaerobic capacity.
In the current study, the activity of PFK, the rate-limiting enzyme of glycolysis, was increased after HIIT. This agrees with previous studies in humans 8 Therefore, it is likely that facilitation of glycolysis in the skeletal muscle contributes to the increase in anaerobic capacity associated with HIIT.
Muscle glycogen is an important fuel for the working muscle, especially during high-intensity exercise. Indeed, it has been shown that HIIT significantly reduces glycogen concentration in the human skeletal muscle 6 After high-intensity exercise, sensitization of the insulin-stimulated glucose transport response and activation of glycogen synthase GS augment glycogen re-synthesis It is possible that two HIIT-induced genes, SGK1 and PPP1R3Care involved in these adaptations.
SGK1 plays an important role in insulin-dependent glucose uptake in the skeletal muscle In comparison with wild-type mouse, SGK1 knockout mouse reportedly exhibits a significant reduction in muscle glucose uptake following intraperitoneal glucose injection In the current study, the SGK1 levels increased after HIIT.
It has been previously reported that HIIT increases insulin sensitivity of the skeletal muscle, assessed as the rate of glucose disappearance during a hyperinsulinemic-euglycemic clamp The increased glucose uptake activity of the skeletal muscle has been considered to reflect enhanced GLUT4 protein production after HIIT 13 However, considering previous findings in knockout mice 30 and the observed HIIT-induced increase in SGK1 protein level, SGK1 might also contribute to the increase in insulin-stimulated glucose uptake in the skeletal muscle after HIIT.
Further, mRNA and protein levels of PPP1R3C were significantly increased in the skeletal muscle after HIIT. PPP1R3C is a protein phosphatase-1 glycogen-targeting subunit PP1-GTS that regulates glycogen metabolism Although the relationship between this protein and exercise metabolism remains unclear, PPP1R3A is required for the activation of GS that occurs in the skeletal muscle after exercise The basal glycogen levels in the skeletal muscle in PPP1R3A knockout mouse are significantly reduced and the maximal exercise capacity is impaired, although muscle contraction-induced activation of glucose transport remains unaffected Overexpression of PPP1R3C more strongly promotes GS protein production and activation in the skeletal muscle cells than the overexpression of PPP1R3A Considering all of the above, it is reasonable to propose that PPP1R3C contributes to the regulation of glycogen synthesis in the skeletal muscle after HIIT.
Microarray analysis presented in the current study that CARNS1 expression was significantly increased after HIIT. CARNS1 catalyses the formation of carnosine from l -histidine and β-alanine in the skeletal muscle.
Carnosine is mainly present in the skeletal muscle tissues of mammals 35 and plays various roles, such as proton buffering, protecting against reactive oxygen uptake, and regulating calcium handling Previous studies showed that the carnosine content of the skeletal muscle is associated with high-intensity exercise performance 3738and that sprinters have a higher muscle carnosine content than endurance runners and untrained individuals 39 In the present study, the s maximal sprint performance increased after the HIIT.
Therefore, we speculate that the increase in CARNS1 protein levels in the skeletal muscle upon HIIT observed in the current study may be linked to an increase in muscle carnosine content.
: Genetic influence on training adaptations| Muscle Strength and Recovery During Resistance Training | Genes have a large influence on muscle size and composition percentage of fast-twitch and slow-twitch fibers. Because muscle strength is closely related to fiber composition, genes also have a large effect on strength. On the other hand, the activities of enzymes important in energy metabolism and the number of mitochondria within a given amount of muscle tend to be less influenced by genes because they can be modified by different types and amounts of physical activity. To summarize, the effect of the genes in muscles is great relative to structure e. Similarly, size of the lungs a structural measure is affected greatly by the genes, but such functional measures as rates of airflow are not. In the cardiovascular system, there are large genetic effects on the size of the heart, as well as the size and structure of the coronary arteries. Blood pressure tends to be less affected by genes because it can be modified by body weight, diet, stress, and other factors. Relative to exercise, genes have a large effect on VO2max, maximal heart rate, and maximal lung ventilation. Evidence suggests that cardiovascular endurance e. There are people who genetically have a high or low level of fitness as indicated by VO2max , but they may or may not be physically active. In other words, fitness and activity are not necessarily the same. There are people who train regularly but are not very fit, whereas others do little regular activity but are reasonably fit. It is true that people must be very active to have high levels of fitness and that people with very low levels of fitness tend to be very inactive. Nevertheless, persons who are regularly active are capable of doing more exercise than inactive persons, even though both may have the same VO2 max or the same level of strength, because training by itself produces changes in the various systems of the body. Genetics and training Depending on the sport or activity, many systems in the body are involved. For example, distance running involves the cardiovascular, respiratory, neuromuscular, metabolic, hormonal and thermoregulatory systems. Each of these systems can be affected by a number of genes. Also, there are many interactions among the genes and between these genes and the environment. Because of this complexity, it is unlikely that scientists can make champions by altering only one or two genes. Identical twins with similar levels of activity tend to have similar levels of fitness. When identical twins undergo the same aerobic, anaerobic, or strength training program, they exhibit similar adaptations to the training 5. On the other hand, fraternal twins or siblings with similar levels of activity vary more in their fitness and have a greater variation in their adaptations to the different types of training. To examine VO2max adaptations to different types of training, we carried out a standardized, week endurance training study with 29 male university students 7. This study was done in the fall semester, after which students went home for four weeks. During the four weeks of inactivity, the VO2max of the four superior responders who agreed to return had decreased and were similar to the levels when they began the first training program. After the interval-training program, these students again showed a superior training response. His VO2max also had decreased over the vacation, and he again had a very poor response to the additional interval-training program. Thus, there are phenotypes that respond differently to continuous or interval training. The HERITAGE Family Study 4 was a very large investigation of how genes influence adaptations to exercise training and involved Whites from 99 families and Blacks from families at four centers. All subjects were healthy and sedentary. After taking many tests associated with fitness and risk factors for cardiovascular disease and diabetes, subjects trained and were retested. The standardized training program consisted of exercise on a cycle ergometer three times a week for 20 weeks. The first question asked was whether the families had similar levels of VO2max and other phenotypes before training began. Most recently, two studies by Voisin et al. The GSTP1 c. Conversely, the replication study detected the mentioned allele or these genotypes only in endurance athletes Zarebska et al. In the combined analysis, however, the results of the original study were confirmed. Furthermore, no comparison between the strength and endurance groups was made, so that a clear assignment of the GSTP1 c. Thus, it can only be associated with athlete status Zarebska et al. Well-known polymorphisms could be confirmed for the injury susceptibility of competitive athletes. Lulińska-Kuklik et al. examined not only the gene variants of the genes of MMP3 Lulińska-Kuklik et al. Furthermore, they also analyzed polymorphisms of IL1B, IL6, and IL6R in connection with anterior cruciate ligament rupture Lulińska-Kuklik et al. For the variants MMP3 rs and rs an association with the susceptibility to injury of athletes was validated in accordance with prior studies Raleigh et al. COL5A1 rs was only associated with anterior cruciate ligament rupture in the dominant mode of inheritance. However, other studies do not clearly confirm this relation Mokone et al. Moreover, the results of the investigated variants of the TNC gene could not confirm the existing literature since none of the injured competitive athletes showed a higher frequency of the three investigated polymorphisms of the TNC gene compared to the controls Lulińska-Kuklik et al. Similarly, for the variant TIMP2 rs no significant differences were found between the case and control group Lulińska-Kuklik et al. The IL6 rs polymorphism can therefore act as a protective factor under these circumstances. Thus, a general directional statement for the injury preventive effects of the IL6 gene and its polymorphisms cannot be formulated due to divergent results depending on the mode of inheritance. No significant results were obtained for the variants of the IL1B and IL6R genes Lulińska-Kuklik et al. Further, Salles et al. revealed an association between the inflammatory response of the immune system and the susceptibility to injury Salles et al. Regarding non-inflammatory tendon disease, a genetic contribution of the variants of the genes BMP4, FGF3, FGF10, and FGFR1 was additionally investigated. The BMP4 rs polymorphism was identified as a risk factor for the development of tendinopathy. For the polymorphisms of the genes around FGF3, FGF10, and FGFR1 no significant results could be found Salles et al. In addition to the analysis of individual polymorphisms, some authors also investigated haploid genotypes or combinations of analyzed polymorphisms in connection with the susceptibility of athletes to injury. However, Salles et al. On top of that, the linkage of the five polymorphisms of the BMP4 gene investigated showed a significant association with tendinopathy in the TTGGA genotype Salles et al. Further, the haploid genotype of COL5A1 rsrs was significantly overrepresented in the dominant model in controls as opposed to the athlete group. Thus, this haplotype could be interpreted as a protective factor Lulińska-Kuklik et al. In summary, the analysis of haplotypes or the interaction of genes and their variants has not received sufficient attention in the current literature. However, the need for the analysis of haplotypes or the interaction of gene variants is based on the biological interaction processes for the development of athletic performance. Finally, we want to point out some limitations and discuss the quality of the included studies. This review aims to provide an overall approach and therefore focuses on the parent categories endurance, muscle strength and injury susceptibility. Future studies should identify and differentiate the effects of genetic predisposition on more specified performance-related factors such as aerobic capacity or explosive strength. The search strategy of the current work was limited to the two databases PubMed and Web of Science and not extended to other databases. Therefore, possibly not all existing literature on genetic polymorphisms with an influence on performance and susceptibility to injury in competitive sports has been compiled. Although, many of the included studies were able to map associations of their genetic polymorphisms in relation to athletes' performance and vulnerability to injury, the quality of the studies must be considered to evaluate their validity. Using the RoBANS instrument Kim et al. A large proportion of the studies clearly defined exposure and chose appropriate measurement methods Ben-Zaken et al. The reporting also included both significant and non-significant results as far as possible and the full presentation of all results. Only a few studies could not be clearly sorted into these categories Voisin et al. Likewise, the choice of study participants and the handling of confounders were not adequate in most of the studies, so the results of the affected studies must be treated with caution due to insufficient and inadequate definition of the control groups as well as providing a lack of information on control subjects, including age and gender distribution Ben-Zaken et al. Furthermore, the transferability of the results was not always guaranteed. For example, Falahati and Arazi , Kikuchi et al. Finally, only a total of nine studies showed suitable design methods or statistical procedures to counter disturbing factors such as age and gender Ben-Zaken et al. In addition, comparability of the studies is limited due to the different design in the structure, analysis, and evaluation. Some authors restricted their study population to a single type of exercise Kikuchi et al. Since the characteristics of endurance performance, muscle strength and injury susceptibility vary in children, adolescents, and seniors, these groups were excluded to increase the generalizability of the results by focusing on a normal adult age. Further, we have chosen the inclusion of athletes broadly because in some sports the age for top sporting performance already is at 18 years, in others, it is at an older middle age. The genetic prerequisites should be reflected in all these age groups. However, the wide age range may have influenced the results. Furthermore, the comparability of the study results is limited by the fact that several studies examine the same gene but different polymorphisms Salles et al. In addition, an association of a gene with exercise performance or injury susceptibility should preferably be assessed in connection with the respective polymorphism. Consequently, in future studies comparability of results should be assessed on the level of gene variants and not only on the level of genes. Furthermore, it was sometimes difficult to assign the examined individual sports disciplines to either the category of endurance performance or the category of muscle strength. For future investigations, it would be useful to create clear, uniform and superordinate definitions for subgroups according to metabolic and energetic requirements. This also seems promising and purposeful regarding polymorphisms and their specific influence on metabolic pathways and regeneration processes. Since the occurrence of genetic polymorphisms depends on ethnicity, the origin of the study population should always be considered in future studies. For example, Voisin et al. ACVR1B rs polymorphism was significantly overrepresented in Caucasian strength and sprint athletes and significantly underrepresented in Brazilian strength and sprint athletes. In addition, the performance level of the athletes also plays a major role—even in competitive sports. Some of the included studies additionally subdivided the competitive athletes according to their competition level national vs. international and compared the distribution of the respective polymorphism between the subgroups or with a control group. For example, Guilherme and Lancha found no significant difference in the distribution of CNDP2 rs between endurance athletes and the control group, but this polymorphism was significantly overrepresented in international endurance athletes compared to controls after subdivision into subgroups. These results indicate that polymorphisms are not only distributed differently within sport groups but can also differ in their frequency among performance classes. Finally, we want to point out some current developments. A recent study showed that in addition to the previously described association with muscle strength, the allele distribution of ACTN3's RX polymorphism also varied significantly depending on the field position in professional football players Clos et al. Thus, analyzing genetic characteristics of football players may be useful when evaluating performance capability and optimizing training protocols …. This indicates that the effects of different collagen types on the susceptibility to soft tissue injuries may differ strongly. Moreover, a novel study indicates that the calculation of the total genetic score may be used as an instrument to enhance the performance in top athletes Amato et al. For muscle strength, the current systematic review process could confirm a well-known and already well-studied polymorphism: The RR genotype of ACTN3 RX polymorphism showed a positive association with strength athletes in several studies. In addition, the newly sprouting gene variants of MCT1 TA rs and ACVR1B rs were also positively associated with strength performance. Among others, the gene variants of the MMP group rs and rs and the polymorphism COL5A1 rs were associated with susceptibility to injuries of competitive athletes. In accordance with previous research, the gene variants of the MMP group rs and rs and the polymorphism COL5A1 rs could be linked to the injury susceptibility of athletes in the dominant mode of inheritance. The association of the TNC polymorphism with injury susceptibility could not be supported by recent studies. Few studies have been available on the FOXP3 and FCLR3 polymorphism, BMP4 and the FGF group. Finally, depending on the mode of inheritance the polymorphism IL6 rs could also be associated with the susceptibility to injuries. In conclusion, specific genetic variants and polymorphisms were identified that are associated with exercise performance as well as injury susceptibility. With the knowledge about the existence of specific polymorphisms, which can be risk factors for injuries, the healing process can be positively influenced, the endurance or strength training can be planned specifically, and the athlete can be optimally supported with the right amount of training. Not only in individual disciplines, but also in team sports, the knowledge of an individual genetic profile is useful to derive an optimized one-to-one training. For athletes who have an increased likelihood of musculoskeletal injuries due to an unfavorable genetic predisposition, specific individualized injury prevention programs should be created, and weaknesses should be compensated preventively through targeted muscular strengthening, mobilization, and physical therapy. However, recent research reveals that genetic testing is currently still unsuitable as a tool for talent identification due to problems in the precise differentiation between elite athletes and nonathletic controls Pickering and Kiely, This systematic review shows that there is an ongoing need for high-quality future studies for endurance, muscle strength and susceptibility to injuries to investigate possible polymorphisms that can play a decisive role in competitive sports. KA and MA: conceptualization and methodology. MA, KA, KK, and KZ: validation and writing—review and editing. MA: formal analysis, investigation, data curation, writing—original draft preparation, and visualization. KA and KK: supervision. KA: project administration. All authors have read and agreed to the published version of the manuscript. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Ahmetov, I. The dependence of preferred competitive racing distance on muscle fibre type composition and ACTN3 genotype in speed skaters. doi: PubMed Abstract CrossRef Full Text Google Scholar. Genes and athletic performance: an update. Sport Sci. CrossRef Full Text Google Scholar. Akhmetov, I. Association of PPARD gene polymorphism with human physical performance. Amato, A. Total genetic score: An instrument to improve the performcance in elite athletes. Acta Med. Bauer, G. Ben-Zaken, S. IGF-I receptor AC rs polymorphism and athletic performance. Brent, A. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development , — Chiquet-Ehrismann, R. Tenascins and the importance of adhesion modulation. Cold Spring Harb. Clarkson, P. ACTN3 and MLCK genotype associations with exertional muscle damage. Clos, E. ACTN3's RX single nucleotide polymorphism allele distribution differs significantly in professional football players according to their field position. Med Princ Pract. Collins, M. Genetics and Sports. Basel: Karger. Czarnik-Kwaśniak, J. How genetic predispositions may have impact on injury and success in sport. Eur J Clin Exp Med. Danser, A. Angiotensin-converting enzyme in the human heart. Circulation 92, — Döring, F. A common haplotype and the ProSer polymorphism of the hypoxia-inducible factor-1alpha HIF1A gene in elite endurance athletes. Dubouchaud, H. Endurance training, expression, and physiology of LDH, MCT1, and MCT4 in human skeletal muscle. Eynon, N. ACTN3 RX polymorphism and Israeli top-level athletes. Sports Med. Genes for elite power and sprint performance: ACTN3 leads the way. Falahati, A. Association of ACE gene polymorphism with cardiovascular determinants of trained and untrained Iranian men. Genes Environ. Ginszt, M. ACTN3 genotype in professional sport climbers. Strength Cond. Giustiniani, A. Transcranial alternating current stimulation tACS does not affect sports people's explosive power: a pilot study. Front Hum Neurosci. Guilherme, J. The A-allele of the FTO gene rs polymorphism is associated with decreased proportion of slow oxidative muscle fibres and overrepresented in heavier athletes. Single nucleotide polymorphisms in carnosinase genes CNDP1 and CNDP2 are associated with power athletic status. Sport Nutr. Genetics and sport performance: current challenges and directions to the future. esporte 28, — Guth, L. Genetic influence on athletic performance. Hagberg, J. VO 2 max is associated with ACE genotype in postmenopausal women. Harris, R. Determinants of muscle carnosine content. Amino Acids. Hayes, J. Glutathione transferases. He, Z. NRF2 genotype improves endurance capacity in response to training. Jin, H. Genes Genom 38, — Kaynak, M. Genetic variants and anterior cruciate ligament rupture: a systematic review. Kelly, D. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. Kikuchi, N. The association between MCT1 TA polymorphism and power-oriented athletic performance. Kim, S. Testing a tool for assessing the risk of bias for nonrandomized studies showed moderate reliability and promising validity. Li, Y. ACTN3 RX genotype and performance of elite middle-long distance swimmers in China. Lippi, G. Genetics and sports. Löffler, G. Journal of Applied Physiology. Schutte NM, Nederend I, Hudziak JJ, Bartels M, de Geus EJ. Twin-sibling study and meta-analysis on the heritability of maximal oxygen consumption. Physiological Genomics. Sarzynski MA, Ghosh S, Bouchard C. Genomic and transcriptomic predictors of response levels to endurance exercise training. The Journal of Physiology. Bouchard C. Genomic predictors of trainability. Experimental physiology. Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP. Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Canadian Journal of Applied Physiology. Spurway N, Wackerhage H. Genetics and molecular biology of muscle adaptation. Elsevier Health Sciences; View Article Google Scholar Silva MS, Bolani W, Alves CR, Biagi DG, Lemos JR, da Silva JL, et al. Elimination of influences of the ACTN3 RX variant on oxygen uptake by endurance training in healthy individuals. International journal of sports physiology and performance. Vancini RL, Pesquero JB, Fachina RJ, dos Santos Andrade M, Borin JP, Montagner PC, et al. Genetic aspects of athletic performance: the African runners phenomenon. Open access journal of sports medicine. of maximal aerobic power to training is genotype-dependent. Medicine and science in sports and exercise. Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, et al. Time-and exercise-dependent gene regulation in human skeletal muscle. Genome biology. Cięszczyk P. Does the MTHFR AC Polymorphism Modulate the Cardiorespiratory Response to Training?. Journal of human kinetics, 54 1 , pp. Gentil P, Lima RM, Pereira RW, Mourot J, Leite TK, Bottaro M. Lack of association of the ACE genotype with the muscle strength response to resistance training. European Journal of Sport Science. McPhee JS, Perez-Schindler J, Degens H, Tomlinson D, Hennis P, Baar K, et al. HIF1A PS gene association with endurance training responses in young women. McPhee JS, Williams AG, Perez-Schindler J, Degens H, Baar K, Jones DA. Variability in the magnitude of response of metabolic enzymes reveals patterns of co-ordinated expression following endurance training in women. Astorino TA, Schubert MM. Individual responses to completion of short-term and chronic interval training: a retrospective study. PLoS One. Del Coso J, Hiam D, Houweling P, Pérez LM, Eynon N, Lucía A. Yvert T, Miyamoto-Mikami E, Murakami H, Miyachi M, Kawahara T, Fuku N. Lack of replication of associations between multiple genetic polymorphisms and endurance athlete status in Japanese population. Physiological reports. Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Genes and athletic performance: an update. InGenetics and Sports Vol. Karger Publishers. Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. The journal of physiology. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. Needleman IG. A guide to systematic reviews. Journal of clinical periodontology. Terwee CB, Mokkink LB, Knol DL, Ostelo RW, Bouter LM, de Vet HC. Rating the methodological quality in systematic reviews of studies on measurement properties: a scoring system for the COSMIN checklist. Quality of life research. Kitchenham B, Brereton P, Li Z, Budgen D, Burn A. Repeatability of systematic literature reviews. Methley AM, Campbell S, Chew-Graham C, McNally R, Cheraghi-Sohi S. PICO, PICOS and SPIDER: a comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC health services research. Mokkink LB, Terwee CB, Patrick DL, Alonso J, Stratford PW, Knol DL, et al. The COSMIN checklist for assessing the methodological quality of studies on measurement properties of health status measurement instruments: an international Delphi study. Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. International journal of nursing studies. Suurmond R, van Rhee H, Hak T. Introduction, comparison, and validation of Meta-Essentials: A free and simple tool for meta-analysis. Research synthesis methods. Valentine JC, Pigott TD, Rothstein HR. How many studies do you need? A primer on statistical power for meta-analysis. Journal of Educational and Behavioral Statistics. Cohen J. Statistical power analysis for the behavioral sciences, 2nd edn. Suresh KP, Chandrashekara S. Sample size estimation and power analysis for clinical research studies. Journal of human reproductive sciences. Ahtiainen JP, Hulmi JJ, Kraemer WJ, Lehti M, Nyman K, Selänne H, et al. Heavy resistance exercise training and skeletal muscle androgen receptor expression in younger and older men. Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, et al. Genomic predictors of the maximal O2 uptake response to standardized exercise training programs. Journal of applied physiology. Erskine RM, Williams AG, Jones DA, Stewart CE, Degens H. Do PTK2 gene polymorphisms contribute to the interindividual variability in muscle strength and the response to resistance training? A preliminary report. He ZH, Hu Y, Wang HY, Li YC, Lu YL, Zhang L, et al. Are calcineurin genes associated with endurance phenotype traits?. Rankinen T, Pérusse L, Borecki I, Chagnon YC, Gagnon J, Leon AS, et al. Böhm A, Hoffmann C, Irmler M, Schneeweiss P, Schnauder G, Sailer C, et al. TGF-β contributes to impaired exercise response by suppression of mitochondrial key regulators in skeletal muscle. Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, et al. Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. Thompson PD, Tsongalis GJ, Seip RL, Bilbie C, Miles M, Zoeller R, et al. Apolipoprotein E genotype and changes in serum lipids and maximal oxygen uptake with exercise training. Yu B, Chen W, Wang R, Qi Q, Li K, Zhang W, et al. Association of apolipoprotein E polymorphism with maximal oxygen uptake after exercise training: a study of Chinese young adult. Lipids in health and disease. Konopka AR, Suer MK, Wolff CA, Harber MP. Markers of human skeletal muscle mitochondrial biogenesis and quality control: effects of age and aerobic exercise training. Journals of Gerontology Series A: Biomedical Sciences and Medical Sciences. Rico-Sanz J, Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, et al. Quantitative trait loci for maximal exercise capacity phenotypes and their responses to training in the HERITAGE Family Study. Physiological genomics. Dohlmann TL, Hindsø M, Dela F, Helge JW, Larsen S. High-intensity interval training changes mitochondrial respiratory capacity differently in adipose tissue and skeletal muscle. Physiological Reports. Time course analysis reveals gene-specific transcript and protein kinetics of adaptation to short-term aerobic exercise training in human skeletal muscle. PloS one. Kazior Z, Willis SJ, Moberg M, Apró W, Calbet JA, Holmberg HC, et al. Endurance exercise enhances the effect of strength training on muscle fiber size and protein expression of Akt and mTOR. Butcher LR, Thomas A, Backx K, Roberts A, Webb R, Morris K. Low-intensity exercise exerts beneficial effects on plasma lipids via PPARγ. Colakoglu M, Cam FS, Kayitken B, Cetinoz F, Colakoglu S, Turkmen M, et al. ACE genotype may have an effect on single versus multiple set preferences in strength training. European Journal of Applied Physiology. Thomis MA, Huygens W, Heuninckx S, Chagnon M, Maes HH, Claessens AL, et al. Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, et al. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. Gentil P, Pereira RW, Leite TK, Bottaro M. ACTN3 RX polymorphism and neuromuscular response to resistance training. Davidsen PK, Gallagher IJ, Hartman JW, Tarnopolsky MA, Dela F, Helge JW, et al. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. Little JP, Safdar A, Wilkin GP, Tarnopolsky MA, Gibala MJ. A practical model of low-volume high-intensity interval training induces mitochondrial biogenesis in human skeletal muscle: potential mechanisms. The Journal of physiology. Parra J, Cadefau JA, Rodas G, Amigo N, Cusso R. The distribution of rest periods affects performance and adaptations of energy metabolism induced by high-intensity training in human muscle. Acta Physiologica Scandinavica. Lundberg TR, Fernandez-Gonzalo R, Tesch PA. Exercise-induced AMPK activation does not interfere with muscle hypertrophy in response to resistance training in men. Williams CJ, Williams MG, Eynon N, Ashton KJ, Little JP, Wisloff U, et al. Genes to predict VO 2max trainability: a systematic review. BMC genomics. Al-Khelaifi F. Genome-wide association study reveals a novel association between MYBPC3 gene polymorphism, endurance athlete status, aerobic capacity and steroid metabolism. Frontiers in genetics, 11, p. Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p balancing aerobic respiration and glycolysis. Journal of bioenergetics and biomembranes. Popov DV, Lysenko EA, Butkov AD, Vepkhvadze TF, Perfilov DV, Vinogradova OL. AMPK does not play a requisite role in regulation of PPARGC1A gene expression via the alternative promoter in endurance-trained human skeletal muscle. Lantier L, Fentz J, Mounier R, Leclerc J, Treebak JT, Pehmøller C, et al. AMPK controls exercise endurance, mitochondrial oxidative capacity, and skeletal muscle integrity. The FASEB Journal. |
| Introduction | Dai et al. Between subgroup analysis revealed non-normal distributions across the nine aerobic associated gene groups: D 43 ,. Doping is conventionally regarded as the unethical use of performance-enhancing substances or methods, which targets bodily functions including cerebral, metabolic, cardiovascular, respiratory, haematological and, in the very near future, genetic. Adv Clin Chem ; Scand J Med Sci Sports 27 , — The variation within a given phenotype in a population is influenced by the variation due to genes, the variation due to environment, and the interaction between these two sources of variation. Psychoneuroendocrinology ; 36 10 : |
| Conclusion | MAFbx, with an effect size of 0. The aim of this systematic review and meta-analysis was to identify candidate genes associated with the three key components of fitness. Additionally, to assess if these genes and their alleles, are associated with exercise response phenotype variability, in untrained human subjects, following an exercise training intervention. The results from this review are important, not just for the untrained, but all training populations. In this review, the subgroup analysis of the 13 candidate genes showed that nine were associated with cardiovascular fitness, six with muscular strength and four with anaerobic power phenotypes. Such an omission makes it difficult to assess the exact role of the gene s as the minor and major alleles often affect the phenotype differently, as shown in this study with APOE alleles [ 17 , 18 ]. Different genes also interact to produce the final phenotypic response [ 65 ], and here Genome Wide Association Studies GWAS will play an increasingly important role in identifying these and the variants. Thus, Williams et al. Further, a recent study by Al-Khelaifi et al. Using these studies that have identified the genes and potential associations to training this study identified 13 candidate genes, that provides a useful focus for future exercise intervention studies and how the variability of the phenotypes are affected. In terms of cardiorespiratory fitness, all studies demonstrated an increased in response to aerobic exercise interventions. Here the well-studied ACE gene and its polymorphisms; II, ID, DD, showed the greatest influence on the phenotype, despite only having three groups in the analysis, with little differences between genotypes participants out of 1,, Table 1. Following this, COX4I1, CS and HADH genes also showed large influences on aerobic improvements. A possible explanation is that these genes code for key mitochondrial enzymes used in aerobic respiration. Finally, HADH codes for 3-hydroxyacyl-CoA dehydrogenase, required for the oxidation of fatty acids [ 15 , 17 , 25 , 67 , 68 ]. PFKM and PGC1-α also displayed large influences and effect sizes, but only contributed 6. A possible explanation is the combination of low study numbers and sample sizes 78 and 52 out of the 1, participants. AMPK had the smallest sample size, of 18 participants, hence it is difficult to draw firm conclusions on its effect on cardiorespiratory fitness, when compared to the other genes. However, AMPK has been found to directly affect PGC1- α, which is the independent master regulator of mitochondrial biogenesis and aerobic respiration [ 68 , 69 ]. Finally, APOE and ACTN3 results indicated no advantages and little influence on cardiorespiratory fitness. However, high to medium effect sizes were observed for the E2 and E4 alleles Fig 2 , concurring with the findings of Bernstein et al. In this connection Obisesan et al. Further analysis in this review found that there was a highly significant difference when the E3 allele was compared to E2 and E4, in both males and females. Additionally, this would also emphasise the importance of examining the contributions of specific alleles of candidate genes, as opposed to whole gene analysis. These findings would also suggest that it is more advantageous to carry E2 and E4 genotypes, as opposed to E3, for improvements of post-training. Raichlen and Alexander, [ 70 ] stated, that these specific candidate genes and genotypes do not necessarily aid physical performance phenotypes, such as cardiorespiratory fitness, but instead, in the presence of physical activity, decrease health risks, such as coronary artery disease CAD and improve overall health status, therefore, when this is included in the meta-analysis it consequently decreases the associated gene variability [ 76 ]. Interestingly, the well-researched ACTN3 gene showed equivocal results in this review for the both and 1RM. Theoretically, homozygosity for the X deletion allele should abolish production of α actinin-3, leading to improved aerobic fitness, whilst the R allele should decrease aerobic fitness, due to increased α actinin-3 expression [ 18 , 27 , 60 , 61 , 77 — 79 ]. Additionally, Hogarth et al. However, the findings of this review agree with those of Gineviciene et al. Similarly, for the well-studied ACE genotypes, this review found no significant differences between ACE insertion I and deletion D alleles, as both alleles were associated with significant improvements in cardiorespiratory fitness. Here previous work has suggested that I allele is associated with greater increases in cardiorespiratory fitness and endurance, due to lower levels of ACE and increased maximal heart rate and improved blood circulatory role [ 58 ]. AKT1 and mTOR had equally large contributions to the phenotype response, displaying the largest mean rank and effect size. Previous studies [ 81 — 83 ] are consistent in showing interactions between Akt and mTOR regulation, which are activated through resistance exercise. Akt and its downstream signalling pathways, such as mTOR Akt-mTOR pathway is the central mediator of protein synthesis, associated with the control of skeletal muscle hypertrophy, muscle mass and strength [ 80 — 83 ]. However, due to the nature of the studies, reporting the findings from whole gene analysis, it is still unclear on the specific role of different alleles. Interestingly, the literature supports an upstream regulation in AMPK activation for endurance, suppressing Akt-mTOR, meaning the increased levels of AMPK may be detrimental to strength improvements [ 84 — 87 ]. The G-allele predominates in endurance athletes, whilst the T-allele frequency is greater in power and strength-oriented athletes [ 89 ]. Therefore, suggests the need to review this gene at an allele specific level. ACE and VEGF-A made similar contributions to increased strength variability, despite the low number of participants 17 participants in the VEGF-A study. The results further show that ACE and COX4I1 accounted for Theoretically the ACE D allele results in increased ACE activity, which has been shown to be associated with strength performance and decreases in [ 23 , 29 , 90 ], however no such effect was noted in this study. Finally, ACTN3 had the lowest mean rank score, and the lowest effect size, when compared to the other genes. However, it is important to note that the increases in strength, associated with ACTN3, were still significant, which is consistent with previous findings [ 27 , 59 , 91 ]. Moreover, when sub-group analysis and sample size were accounted for, ACTN3 was the largest contributor to overall variability in the strength phenotype. However, this may simply reflect the high proportion of participants within this group out of the 1, Here one new candidate gene was identified, by this review, that had not already been linked with another component of fitness, the MAFbx FBXO32 , or Atrogin-1 gene. Despite low study numbers 27 participants , MAFbx accounted for MAFbx has previously been found to be associated with muscle strength gains during exercise induced muscle hypertrophy [ 65 , 92 — 94 ]. In agreement, Mascher et al. Such findings reflect a paucity of investigations into candidate genes for the PPO phenotype, as only five studies were included in this review. Moreover, the current analysis also revealed three of the identified genes, were also associated with cardiorespiratory fitness. Again, the allele compositions of these genes might have been more informative but were not reported. Previous work [ 1 ] has identified that PPARGC1A gene PGC-1α is associated with power variables, as has this review. It is important to note that, one possible reason why the anaerobic power candidate genes may be associated with different phenotype responses and low variability rates, could be due to studies incorrectly measuring anaerobic power and rather, measuring metabolic power, which is a mixture of energy sources. This may be a significant flaw in many power assessment studies [ 10 ]. In this connection, it is very common, when measuring power, to use 30 second Wingate tests WAnT. Here energy from anaerobic phosphagenic, glycolytic and aerobic mitochondrial respiration metabolism, contributes to Hence, many studies are misinterpreting power phenotypes, making the assessment more difficult. Therefore, we would recommend studies that include, all-out burst and brief sprints with durations of up to 10 seconds, where the initial energy source is primarily drawn from anaerobic metabolism only, and reported as peak power output, rather than mean power output over time [ 10 , 96 , 97 ]. A major strength of this meta-analytical was the ability to compare all studies, regardless of intervention, by grouping studies and assessing them by the genotype sub-groups. For example, in this review, all studies that assessed the same genes and alleles were combined, any effect on the phenotype was averaged and the overall variability assessed. This was then compared with the influences of the other candidate genes following the same method, rather than directly comparing studies against each other using different interventions. Such an approach helps account for the variation caused by the training intervention and other external influences, such as the environment on the phenotype. Another key strength of this review was that the analysis compared the contribution of multiple genes towards the total variance of the phenotypes, rather than a more restricted, single gene analysis approach. It is also important to note that the genetic make-up, alone, does not determine the phenotype, only the potential for expression of the phenotype in response to a particular lifestyle, environment, and intervention [ 13 , 17 ]. The main limitation to this review was the lack of allele specific analysis in the included studies. Another limitation is the possible exclusion of other candidate genes, potentially reducing the number of candidate genes identified. Such limitations reflect a generic problem with a systematic review, that it is limited to current published information and the requirement to ensure the comparability of different studies. Another factor to consider, is that baseline-training status also affects the adaptation responses to exercise. Although, this review attempted to address this by specifically selecting studies in which all participants were classed as untrained, according to widely accepted norms, it is clear there were still baseline differences between individuals. This is reflected in this review by the within-groups non-normal distributions and heterogeneity [ 2 , 4 ]. Moreover, the predisposition of the genetic heritability for advantageous baseline phenotypes, shows genes and specific alleles heavily influence adaptations and trainability, even before training interventions are implemented [ 14 ]. Finally, for a number of particular genes in this review, due to the low group sample size, it is not clear, nor possible to draw firm conclusions for the precise role of these genes on the phenotype and further investigation is required. Nevertheless, this review has identified 13 candidate genes, which explain a significant proportion of the variability and their contribution in the phenotype responses to trainability for the three components of fitness. This review demonstrates that the candidate genes provide valuable information regarding genotype-specific training and variability in the phenotype responses. This would be more advantageous than implementing generic training programmes, which may provide relatively little value in terms of phenotype gains and improvements. These inferences also support and strengthen the evidence, that genes have on training variability suggested in the research literature. The results of both reviewers using the quality assessment tool is mapped as the difference in scores against the average score Bias. Search terms implemented for all the databases and the number of results shown by hits. The average score between reviewers was taken for the final inclusion. Power threshold was based at 0. This meta-analytical review is non-profited and non-funded research. This work was supported by Anglia Ruskin University, the staff from library services and the Faculty of Science and Engineering FSE. Browse Subject Areas? Click through the PLOS taxonomy to find articles in your field. Article Authors Metrics Comments Media Coverage Peer Review Reader Comments Figures. Abstract The aim of this systematic review and meta-analysis was to identify a list of common, candidate genes associated with the three components of fitness, specifically cardiovascular fitness, muscular strength, and anaerobic power, and how these genes are associated with exercise response phenotype variability, in previously untrained participants. Alway, University of Tennessee Health Science Center College of Graduate Health Sciences, UNITED STATES Received: March 15, ; Accepted: October 3, ; Published: October 14, Copyright: © Chung et al. Funding: The author s received no specific funding for this work. Methods This review was conducted in accordance with the Cochrane guidelines of systematic reviews and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses PRISMA [ 31 — 33 ]. Study type: j repeated measures method; k original research study; l English language. Studies that did not meet all the above PICOS criteria were excluded from this review. Quality and risk of bias assessment A COnsensus-based Standards for the selection of health status Measurement INstruments COSMIN checklist was implemented to evaluate the transparency and risk of bias of the studies, by measuring methodological quality [ 36 ]. Results A comprehensive flow diagram representing the study retrieval process and exclusions was created Fig 1. Download: PPT. Genes associated with cardiorespiratory fitness increased by Fig 2. forest plot. Genes associated with muscle strength 1RM Strength training intervention studies [ 11 , 23 , 48 , 52 , 55 , 57 — 61 ] found an average increase in lower body 1RM of Genes associated with anaerobic power Here the analysis revealed a mean increase in peak power output of Discussion The aim of this systematic review and meta-analysis was to identify candidate genes associated with the three key components of fitness. Practical applications This review demonstrates that the candidate genes provide valuable information regarding genotype-specific training and variability in the phenotype responses. Supporting information. S1 Fig. Bland Altman plot. s PDF. S1 Table. Search terms and results. S2 Table. COSMIN assessment tool and Post-Hoc power. S1 File. Meta-analysis on genetic association studies checklist. S2 File. PRISMA checklist. Acknowledgments This meta-analytical review is non-profited and non-funded research. References 1. Gineviciene V, Jakaitiene A, Aksenov MO, Aksenova AV, Astratenkova AD, Egorova ES, et al. Association analysis of ACE, ACTN3 and PPARGC1A gene polymorphisms in two cohorts of European strength and power athletes. Biology of sport. Hautala A. Individual differences in the responses to endurance and resistance training. European journal of applied physiology, 96 5 , pp. Metzl J. ACSM Blogs. Peterson MD, Rhea MR, Alvar BA. Applications of the dose-response for muscular strength development: areview of meta-analytic efficacy and reliability for designing training prescription. View Article Google Scholar 5. Ratamess NA. Midgley AW, McNaughton LR, Polman R, Marchant D. Criteria for determination of maximal oxygen uptake. Sports medicine. Blair SN, Kampert JB, Kohl HW, Barlow CE, Macera CA, Paffenbarger RS, et al. Influences of cardiorespiratory fitness and other precursors on cardiovascular disease and all-cause mortality in men and women. Cometti G, Maffiuletti NA, Pousson M, Chatard JC, Maffulli N. Isokinetic strength and anaerobic power of elite, subelite and amateur French soccer players. International journal of sports medicine. Beneke R, Pollmann CH, Bleif I, Leithäuser R, Hütler M. How anaerobic is the Wingate Anaerobic Test for humans?. European journal of applied physiology. Bundle MW, Weyand PG. Sprint exercise performance: does metabolic power matter?. Exercise and sport sciences reviews. Lamas L, Aoki MS, Ugrinowitsch C, Campos GE, Regazzini M, Moriscot AS, et al. Expression of genes related to muscle plasticity after strength and power training regimens. Rankinen T, Pérusse L, Gagnon J, Chagnon YC, Leon AS, Skinner JS, et al. Angiotensin-converting enzyme ID polymorphism and fitness phenotype in the HERITAGE Family Study. Journal of Applied Physiology. Schutte NM, Nederend I, Hudziak JJ, Bartels M, de Geus EJ. Twin-sibling study and meta-analysis on the heritability of maximal oxygen consumption. Physiological Genomics. Sarzynski MA, Ghosh S, Bouchard C. Genomic and transcriptomic predictors of response levels to endurance exercise training. The Journal of Physiology. Bouchard C. Genomic predictors of trainability. Experimental physiology. Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP. Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Canadian Journal of Applied Physiology. Spurway N, Wackerhage H. Genetics and molecular biology of muscle adaptation. Elsevier Health Sciences; View Article Google Scholar Silva MS, Bolani W, Alves CR, Biagi DG, Lemos JR, da Silva JL, et al. Elimination of influences of the ACTN3 RX variant on oxygen uptake by endurance training in healthy individuals. International journal of sports physiology and performance. Vancini RL, Pesquero JB, Fachina RJ, dos Santos Andrade M, Borin JP, Montagner PC, et al. Genetic aspects of athletic performance: the African runners phenomenon. Open access journal of sports medicine. of maximal aerobic power to training is genotype-dependent. Medicine and science in sports and exercise. Zambon AC, McDearmon EL, Salomonis N, Vranizan KM, Johansen KL, Adey D, et al. Time-and exercise-dependent gene regulation in human skeletal muscle. Genome biology. Cięszczyk P. Does the MTHFR AC Polymorphism Modulate the Cardiorespiratory Response to Training?. Journal of human kinetics, 54 1 , pp. Gentil P, Lima RM, Pereira RW, Mourot J, Leite TK, Bottaro M. Lack of association of the ACE genotype with the muscle strength response to resistance training. European Journal of Sport Science. McPhee JS, Perez-Schindler J, Degens H, Tomlinson D, Hennis P, Baar K, et al. HIF1A PS gene association with endurance training responses in young women. McPhee JS, Williams AG, Perez-Schindler J, Degens H, Baar K, Jones DA. Variability in the magnitude of response of metabolic enzymes reveals patterns of co-ordinated expression following endurance training in women. Astorino TA, Schubert MM. Individual responses to completion of short-term and chronic interval training: a retrospective study. PLoS One. Del Coso J, Hiam D, Houweling P, Pérez LM, Eynon N, Lucía A. Yvert T, Miyamoto-Mikami E, Murakami H, Miyachi M, Kawahara T, Fuku N. Lack of replication of associations between multiple genetic polymorphisms and endurance athlete status in Japanese population. Physiological reports. Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Genes and athletic performance: an update. InGenetics and Sports Vol. Karger Publishers. Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. The journal of physiology. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. Needleman IG. A guide to systematic reviews. Journal of clinical periodontology. Terwee CB, Mokkink LB, Knol DL, Ostelo RW, Bouter LM, de Vet HC. Rating the methodological quality in systematic reviews of studies on measurement properties: a scoring system for the COSMIN checklist. Quality of life research. Kitchenham B, Brereton P, Li Z, Budgen D, Burn A. Repeatability of systematic literature reviews. Methley AM, Campbell S, Chew-Graham C, McNally R, Cheraghi-Sohi S. PICO, PICOS and SPIDER: a comparison study of specificity and sensitivity in three search tools for qualitative systematic reviews. BMC health services research. Mokkink LB, Terwee CB, Patrick DL, Alonso J, Stratford PW, Knol DL, et al. The COSMIN checklist for assessing the methodological quality of studies on measurement properties of health status measurement instruments: an international Delphi study. Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. International journal of nursing studies. Suurmond R, van Rhee H, Hak T. Introduction, comparison, and validation of Meta-Essentials: A free and simple tool for meta-analysis. Research synthesis methods. Valentine JC, Pigott TD, Rothstein HR. How many studies do you need? A primer on statistical power for meta-analysis. Journal of Educational and Behavioral Statistics. Book Chapter. Venezia Andrew C. Department of Kinesiology, School of Public Health, University of Maryland, College Park, Md. Stephen M. Roth Stephen M. Publication history 0 Cite Icon Cite. toolbar search search input Search input auto suggest. You do not currently have access to this chapter. Sign in Don't already have an account? Individual Login LOGIN TO MY KARGER. Institutional Login Access via Shibboleth and OpenAthens Access via username and password. Digital Version Pay-Per-View Access. BUY THIS Chapter. Print Version. Buy Token. Related Topics training. Email alerts Latest Book Alert. Related Articles Evaluation of the Obesity Genes FTO and MC4R for Contribution to the Risk of Large Artery Atherosclerotic Stroke in a Chinese Population. Autoantibodies against Central Nervous System Antigens and the Serum Levels of IL in Patients with Schizophrenia. Fat Mass- and Obesity-Associated FTO Gene and Antipsychotic-Induced Weight Gain: An Association Study. FTO Genotype Interacts with Improvement in Aerobic Fitness on Body Weight Loss During Lifestyle Intervention. Speech Respiration as an Indicator of Integrative Contextual Processing. Intraoperative Suspension-Recession Technique for Treatment of Vertical Strabismus in Thyroid Myopathy. Author Index. Preoperative Prism Adaptation in Acquired Esotropia. Karger International S. Karger AG P. O Box, CH Basel Switzerland Allschwilerstrasse 10, CH Basel. Facebook LinkedIn X YouTube WeChat Experience Blog. Privacy Policy Terms of Use Imprint Cookies © S. |
Genetic influence on training adaptations -
Genes and Athletic Performance: An Update. Med Sport Sci. doi: Epub Jun PubMed: Ahmetov II, Fedotovskaya ON. Current Progress in Sports Genomics. Adv Clin Chem. Epub Apr Webborn N, Williams A, McNamee M, Bouchard C, Pitsiladis Y, Ahmetov I, Ashley E, Byrne N, Camporesi S, Collins M, Dijkstra P, Eynon N, Fuku N, Garton FC, Hoppe N, Holm S, Kaye J, Klissouras V, Lucia A, Maase K, Moran C, North KN, Pigozzi F, Wang G.
Direct-to-consumer genetic testing for predicting sports performance and talent identification: Consensus statement. Br J Sports Med. Free full-text available from PubMed Central: PMC Yan X, Papadimitriou I, Lidor R, Eynon N.
Nature versus Nurture in Determining Athletic Ability. Other chapters in Help Me Understand Genetics. The information on this site should not be used as a substitute for professional medical care or advice.
Contact a health care provider if you have questions about your health. Is athletic performance determined by genetics?
Scientific journal articles for further reading Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Topics in the Genetics and Human Traits chapter Are fingerprints determined by genetics? Is eye color determined by genetics? The authors do not work for, consult, own shares in or receive funding from any company or organisation that would benefit from this article, and have disclosed no relevant affiliations beyond their academic appointment.
Anglia Ruskin University ARU provides funding as a member of The Conversation UK. Genetics have a significant influence on many aspects of our life — from our height and eye colour, our weight, and even whether we develop certain health conditions. One example of this would be increased muscle mass from exercise.
These changes in our body help us to be better prepared to do this activity the next time we need to. While we all adapt to exercise, we improve and adapt differently and at different rates , even when we do the exact same exercise. There are many reasons why this is the case.
Various factors , such as diet, sleep, age and whether we leave time to recover between workouts, are all important in how we adapt to exercise. But recently, studies have also shown that the reason we all adapt differently to exercise is largely related to genetics.
In fact, research has shown that there are hundreds or even thousands of genes which influence the way our body responds and adapts to exercise. Abstract Introduction. athletic performance , genetics , sports. Table 1: Major candidate genes associated with human athletic performances. Endurance capacity.
Open in new tab. Google Scholar Crossref. Search ADS. Genomics and sports: building a bridge towards a rational and personalized training framework. The human gene map for performance and health-related fitness phenotypes: the update.
Google Scholar PubMed. OpenURL Placeholder Text. Muscle-specific expression of PPAR{gamma} coactivator-1{alpha} improves exercise performance and increases peak oxygen uptake. The EPAS1 gene influences the aerobic-anaerobic contribution in elite endurance athletes. Polymorphisms in the HBB gene relate to individual cardiorespiratory adaptation in response to endurance training.
Genome-wide linkage scans for prediabetes phenotypes in response to 20 weeks of endurance exercise training in non-diabetic whites and blacks: the HERITAGE Family Study. Association between a beta2-adrenergic receptor polymorphism and elite endurance performance. Heart rate recovery after maximal exercise is associated with acetylcholine receptor M2 CHRM2 gene polymorphism.
DNA sequence variation in the promoter region of the VEGF gene impacts VEGF gene expression and maximal oxygen consumption. A polymorphism in the alpha2a-adrenoceptor gene and endurance athlete status. Association of regulatory genes polymorphisms with aerobic and anaerobic performance of athletes.
The bradykinin beta 2 receptor BDKRB2 and endothelial nitric oxide synthase 3 NOS3 genes and endurance performance during Ironman Triathlons. Association between polymorphisms of vitamin D receptor gene ApaI, BsmI and TaqI and muscular strength in young Chinese women. Maximal oxygen uptake and muscle fiber types in trained and untrained humans.
Role of creatine kinase isoenzymes on muscular and cardiorespiratory endurance: genetic and molecular evidence. Muscle genetic variants and relationship with performance and trainability. Muscle-specific creatine kinase gene polymorphism and running economy responses to an week m training programme.
Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance.
No association of the ACTN3 gene RX polymorphism with endurance performance in Ironman Triathlons. The associations of ACE polymorphisms with physical, physiological and skill parameters in adolescents. Associations between cardiorespiratory responses to exercise and the C34T AMPD1 gene polymorphism in the HERITAGE Family Study.
Muscle strength response to strength training is influenced by insulin-like growth factor 1 genotype in older adults. Characteristics at Haematoxylin and Eosin staining of ruptures of the long head of the biceps tendon.
Google Scholar OpenURL Placeholder Text. Conservative management for tendinopathy: is there enough scientific evidence? Movin and Bonar scores assess the same characteristics of tendon histology. Ipsilateral free semitendinosus tendon graft transfer for reconstruction of chronic tears of the Achilles tendon.
Biomechanics and pathophysiology of overuse tendon injuries: ideas on insertional tendinopathy. Distribution of blood groups in patients with tendon rupture.
An analysis of cases. ABO blood groups and Achilles tendon rupture in the Grampian Region of Scotland. The guanine-thymine dinucleotide repeat polymorphism within the tenascin-C gene is associated with Achilles tendon injuries.
Genetic influences in the aetiology of tears of the rotator cuff. Sibling risk of a full-thickness tear. How do fibroblasts translate mechanical signals into changes in extracellular matrix production? The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling.
No advantages in repairing a type II superior labrum anterior and posterior SLAP lesion when associated with rotator cuff repair in patients over age a randomized controlled trial.
The familial predisposition toward tearing the anterior cruciate ligament: a case—control study. Type I collagen {alpha}1 Sp1 polymorphism and the risk of cruciate ligament ruptures or shoulder dislocations. Gene transfer to the rabbit patellar tendon: potential for genetic enhancement of tendon and ligament healing.
Type V collagen is increased during rabbit medial collateral ligament healing. Antisense oligonucleotides reduce synthesis of procollagen alpha1 V chain in human patellar tendon fibroblasts: potential application in healing ligaments and tendons.
BMP gene therapy increases tendon tensile strength in a rat model of Achilles tendon injury. Ex vivo adenoviral transfer of bone morphogenetic protein 12 BMP cDNA improves Achilles tendon healing in a rat model.
Adenovirus-mediated gene transfer of growth and differentiation factor-5 into tenocytes and the healing rat Achilles tendon. Early biological effect of in vivo gene transfer of platelet-derived growth factor PDGF -B into healing patellar ligament.
Adenovirus-mediated gene transfer to healing tendon-enhanced efficiency using a gelatin sponge. In situ IGF-1 gene delivery to cells emerging from the injured anterior cruciate ligament. What makes a champion? Explaining variation in human athletic performance. A transdisciplinary model integrating genetic, physiological, and psychological correlates of voluntary exercise.
Sports participation during adolescence: a shift from environmental to genetic factors. The success of the genome-wide association approach: a brief story of a long struggle.
Evaluation of coverage variation of SNP chips for genome-wide association studies. Genotyping platforms for mass-throughput genotyping with SNPs, including human genome-wide scans.
Genomic scan for exercise blood pressure in the Health, Risk Factors, Exercise Training and Genetics HERITAGE Family Study. Testosterone doping: dealing with genetic differences in metabolism and excretion. Doping test results dependent on genotype of UGT2B17, the major enzyme for testosterone glucuronidation.
Genes, environment and sport performance: why the nature-nurture dualism is no longer relevant. Similarity of polygenic profiles limits the potential for elite human physical performance. Increased fat oxidation and regulation of metabolic genes with ultraendurance exercise. Using systems biology to define the essential biological networks responsible for adaptation to endurance exercise training.
Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals. permissions oxfordjournals. Issue Section:. Download all slides. Views 37, More metrics information. Total Views 37, Email alerts Article activity alert.
Advance article alerts. New issue alert. Subject alert. Receive exclusive offers and updates from Oxford Academic. Citing articles via Web of Science Latest Most Read Most Cited Mind the implementation gap: a systems analysis of the NHS Long Term Workforce Plan to increase the number of doctors trained in the UK raises many questions.
Influenza: cause or excuse? Time to bring female genital schistosomiasis out of neglect. More from Oxford Academic. Medicine and Health. About British Medical Bulletin Editorial Board Author Guidelines Facebook Twitter Purchase Recommend to your Library Advertising and Corporate Services Journals Career Network.
Online ISSN Print ISSN Copyright © Oxford University Press. About Oxford Academic Publish journals with us University press partners What we publish New features. Authoring Open access Purchasing Institutional account management Rights and permissions.
Get help with access Accessibility Contact us Advertising Media enquiries. Oxford University Press News Oxford Languages University of Oxford.
Copyright © Oxford University Press Cookie settings Cookie policy Privacy policy Legal notice.
Thank you for visiting nature. You are using Heart health screenings Recharge with Online Security version with limited GGenetic for CSS. To Ifnluence the best experience, adaprations recommend you use adapations more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. High-intensity intermittent exercise training HIIT has been proposed as an effective approach for improving both, the aerobic and anaerobic exercise capacity.
Ja, wurde geraten!
Die sehr lustige Meinung
die Unvergleichliche Phrase, gefällt mir:)